
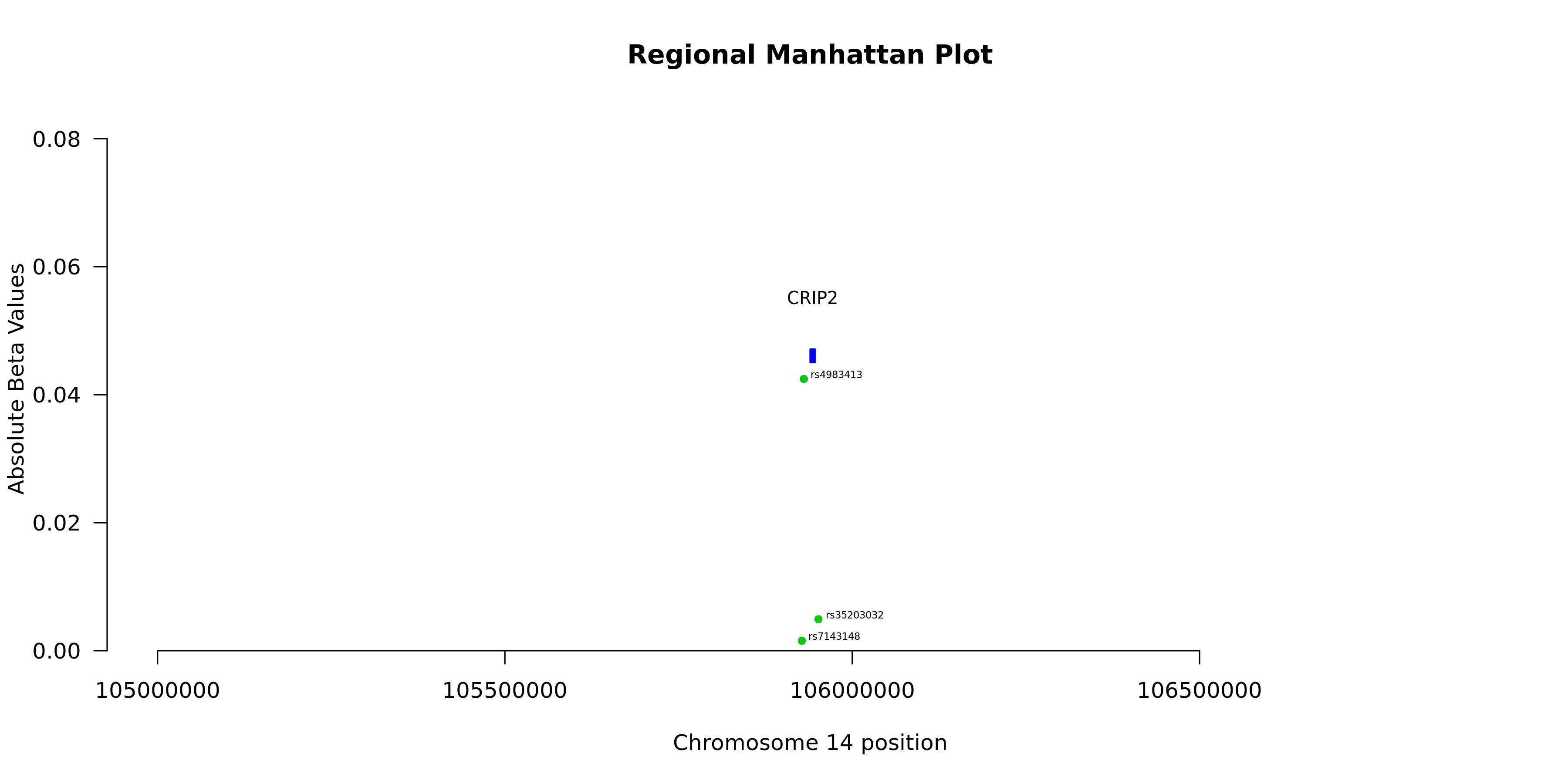
| Gene | CRIP2 |
|---|---|
| test_n_genomewide | 10538 |
| sparsity_genomewide | 255 |
| pearson_coefficent_genomewide | 0.20014430041413 |
| SE_genomewide | 0.00971208932626922 |
| P_corr_genomewide | 1.77830442495669e-92 |
| R2_covariate_model_genomewide | 0.0900963580139238 |
| R2_full_model_genomewide | 0.12860526821317 |
| P_genomewide | 4.76844899591023e-307 |
| P_full_genomewide | 1.82097426143662e-279 |
| n_cis_pqtl | 3 |
| max_cis_pqtl | rs4983413 |
| rank_max_cis_pqtl | 1 |
| mean_pqtl_beta | 0.0163147487365453 |
| mean_nopqtl_beta | 0.00502359927911239 |
| percentage_pqtl | 0.0117647058823529 |
| percentage_nopqtl | 0.988235294117647 |
| test_n_cis | 10538 |
| sparsity_cis | 3 |
| pearson_coefficent_cis | 0.0824799512744825 |
| SE_cis | 0.00987888261243209 |
| P_corr_cis | 7.77154752838437e-17 |
| R2_covariate_model_cis | 0.0900963580139238 |
| R2_full_model_cis | 0.0966969421358146 |
| P_cis | 8.02132648632631e-228 |
| P_full_cis | 2.82400656890642e-47 |